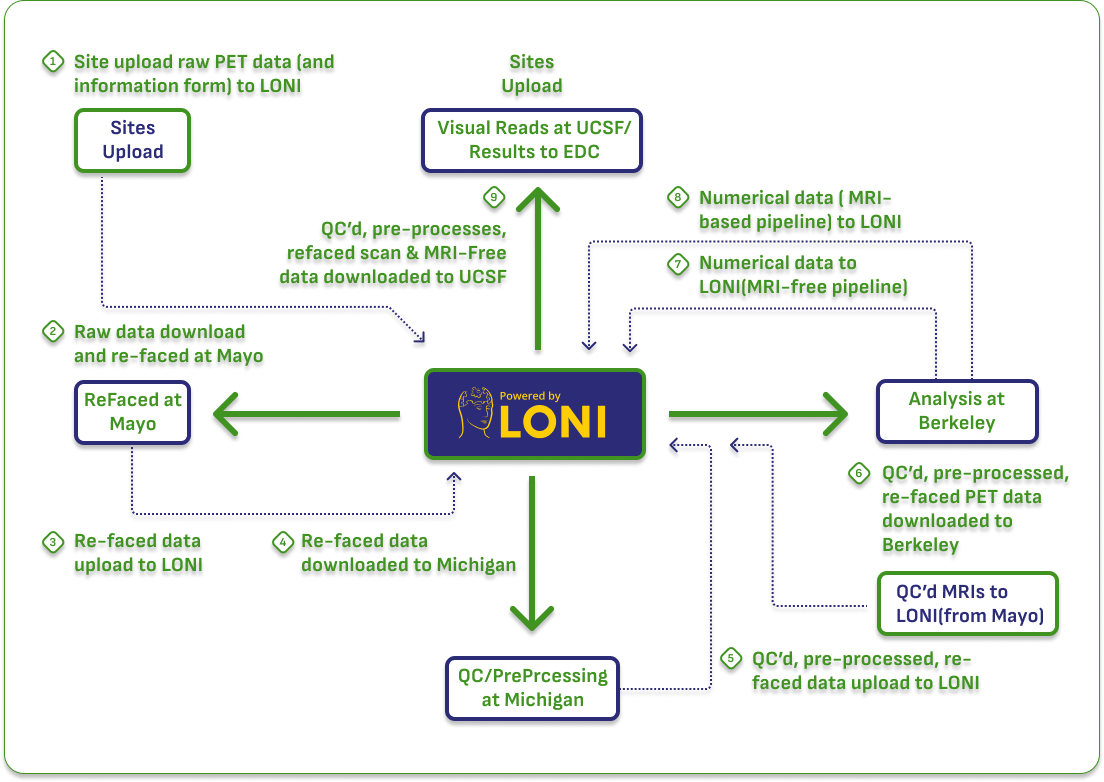
Historical PET Availability and Data Types
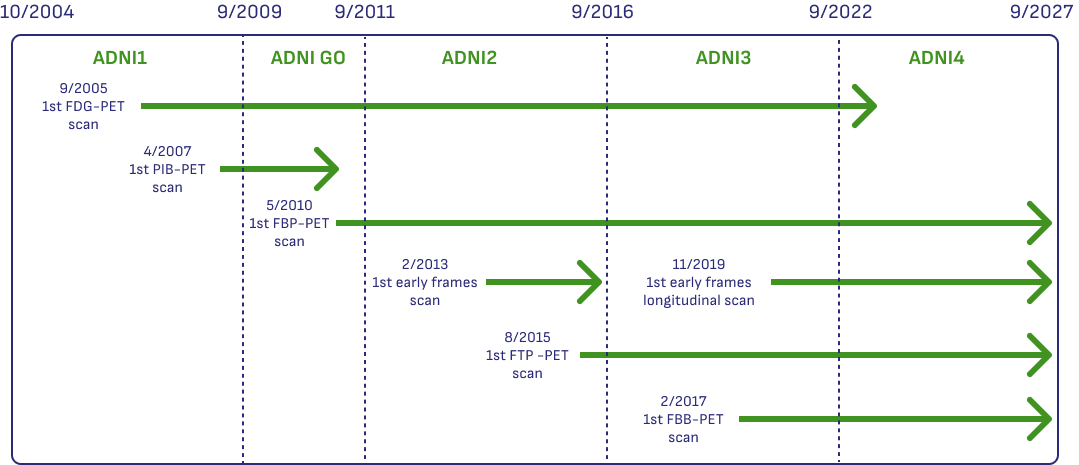
Understanding what kind of PET data is available on a more granular level can be challenging. ADNI has been active for 20 years, and the types of PET data collected has evolved over time.
This graphic shows which scans were collected at different times across the different phases of ADNI, and which PET tracers were used during different periods.
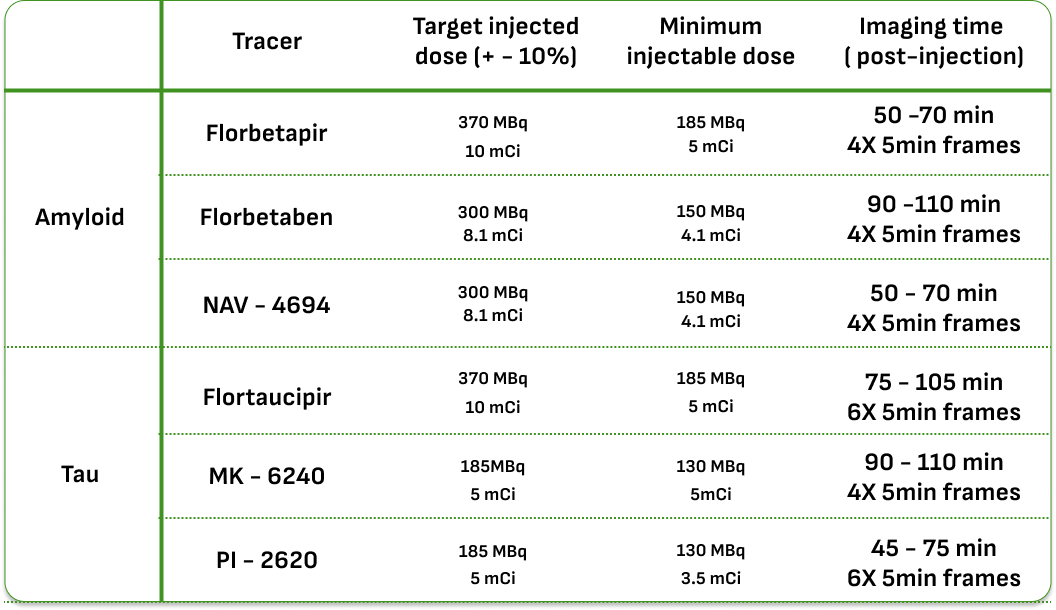
During each phase of ADNI, different participant groups were studied at different intervals. The following table provides this information: