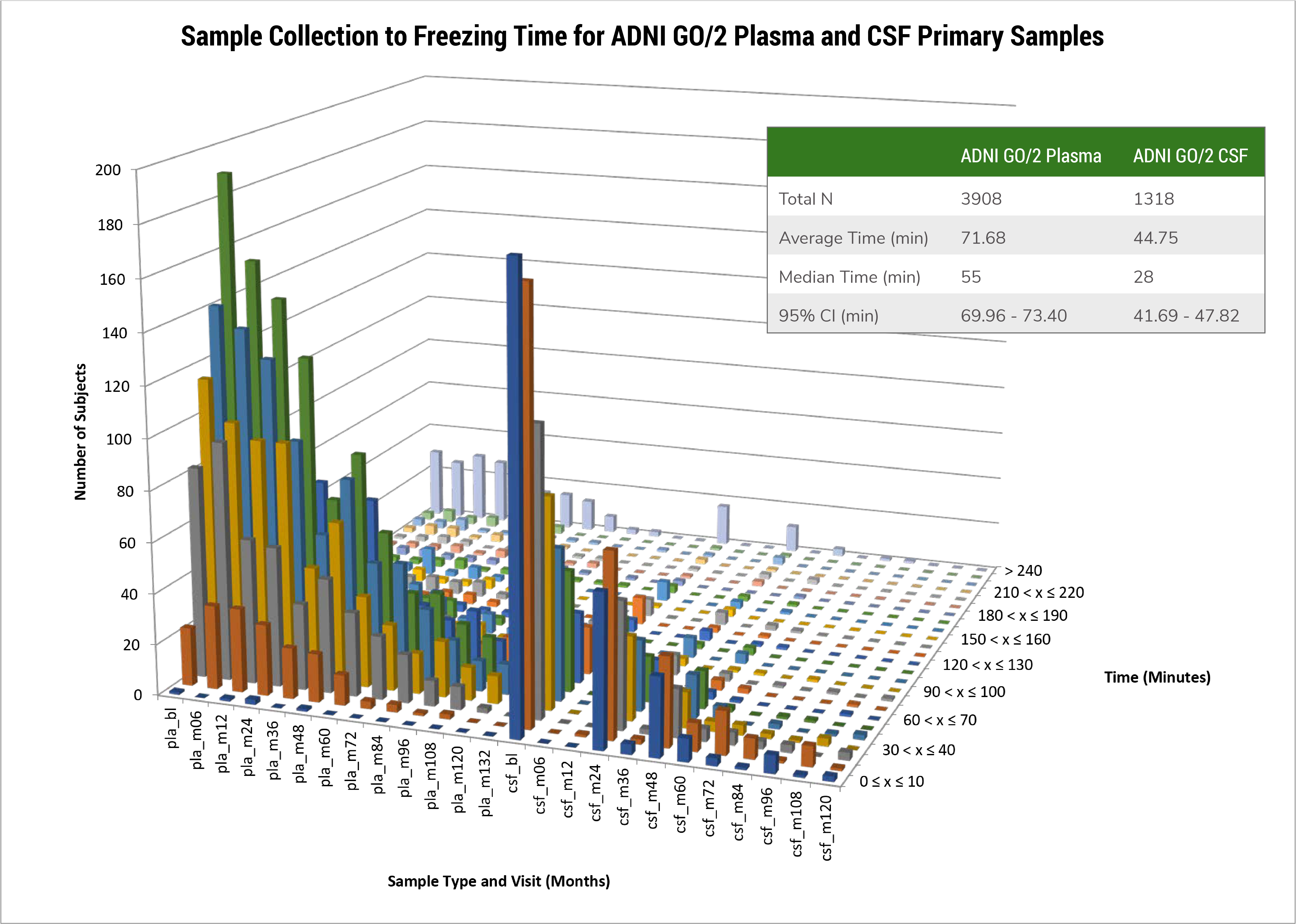
The Biomarker Core in ADNI3 is focusing on 4 areas of activity and studies including: biofluid banking (CSF, plasma and serum) management and pre-analytical assessments; standardization of CSF Ab42, Ab40, t-tau and p-tau181 measurement in ADNI patients using the highly validated Roche Elecsys cobas e 601 fully automated immunoassay platform and reference LC/MSMS methodology for CSF Ab42, Ab40 and Ab38; determination of cut-points for Ab42, t-tau, p-tau181, and ratios using several approaches including ROC analyses using FBP amyloid PET imaging for disease detection and disease independent mixture modeling; collaborative studies on new biomarker development/validation/testing in CSF or plasma by immunoassay or mrm LC/MSMS including NFL, total and phospho-a-synuclein, Vilip-1, sTREM2, progranulin, TDP-43, metabolomics/lipidomic biomarkers and proteomic quantitative assays.
Leslie M Shaw and John Q Trojanowski co-lead the ADNI Biomarker Core at the University of Pennsylvania.
Multicenter Multivariate Studies
Alzheimer’s Disease (AD) Neuroimaging Initiative (ADNI) Related Biofluid And Genetic Biomarker Studies Using Non-ADNI University Of Pennsylvania (UPenn) Biosamples From Patients Followed Longitudinally Affected By AD, Parkinson’s Disease With (PDD) And Without (PD) Dementia, Frontotemporal Degeneration (FTD), Amyotrophic Lateral Sclerosis (ALS) Or Related Neurodegenerative Diseases (ND)
John Q. Trojanowski and Leslie M. Shaw
The Center for Neurodegenerative Disease (ND) Research (CNDR; (http://www.med.upenn.edu/cndr/) is the home base of of several large programmatic grants focusing on basic science and patient oriented research begining in 1990 with an NIA funded PO1 on shared mechanisms of AD and PD led by John Trojanowski (PO1 AG-09215) that ran from 1990 to 2010 and then was continued from 2007 to the present as the Penn Udall Center (P50 NS-053488). In addition, there is a Penn AD Core Center (ADCC; P30 AG-10124-27) led by John Trojanowski, which has a dementia with Lewy Body (DLB) and frontotemporal degeneration (FTD) module, and an NIA funded PO1 on FTD led by Virginia Lee (P01 AG-17586-16) as well as a previously funded PO1 on ALS and TDP-43 (P01 AG032953) led by Virginia Lee. Of course there many other AD and related dementia (ADRD) grants at Penn including the AD Genetics Consortium led by Jerry Schelenberg and Li-San Wang, but the grants listed above follow patients longitudinally.
Thus, CNDR has systematically collected patient biosamples including CSF, DNA/RNA, plasma, serum, whole blood and postmortem brain samples since 1990 from longitudinally followed patients, many of whom consent to autopsy. Accordingly, CNDR has banked CNS tissues from >1,700 subjects with a primary diagnosis of AD, Lewy body demenias (LBD), FTD, ALS, related NDs and normal controls (NC). This includes cases with the following primary neuropathology diagnoses, although many have other or additional ND co-pathologies1 such as aging related astroglial tauopathy (ARTAG):2-4 576 AD, 120 PD, 134 PDD, 52 DLB, 55 MSA brains and 257 NC or pathological aging related tauopathy (PART).5 There also are 111 frontotemporal lobar degeneration (FTLD) due to TDP-32 inclusions (FTLD-TDP), 52 ALS, 50 CBD,106 PSP and 23 PiD brains. DNA from brain is available on >1500 (89%) of autopsies. Some autopsy cases have paired DNA from blood or saliva and brain (n=515). DNA from peripheral blood is banked from approximately 480 NC and 3700 individuals in the following diagnostic categories: 1478 AD, 613 ALS, 573 FTLD, 1003 PD/PDD/DLB, 22 MSA. For archival biofluids, we have CSF samples from >1500 individuals in the following diagnostic categories: 634 AD, 507 FTLD, 202 PD/PDD/DLB, 151 ALS, and 105 NC. We have plasma samples from >3300 unique individuals in the following diagnostic categories: 859 AD, 504 FTLD, 948 PD/PDD/DLB, 545 ALS, and 454 NC. In nearly all cases, CSF is matched with plasma collected at the same time, and for many individuals, multiple samples have been collected longitudinally. Thus, our archived DNA/RNA and biofluid biosamples are extremely valuable for comparative studies across diverse ND and they have been used extensively in neuropathology, genetic and biomarker studies supported by the NIH funded grants noted above.1, 6, 7 Below are brief highlights of genetic and biofluid biomarker studies conducted on Penn non-ADNI biosamples mainly since 2013 that complement the biomarker studies done in ADNI with ADNI patient samples.
Studies conducted using these UPenn samples often include biosamples from other centers or investigators beyond Penn and the focus here is on biomarkers (mainly CSF, plasma and DNA, but some are combined with PET imaging and structural imaging data) and neuropathology. This is exemplified in particular by comparative studiesof AD and PD including: 1) Recognition of the contribution of co-morbid AD and alpha-synuclein (aSyn) pathology to cognitive decline in AD with concomitant aSyn pathlogy and LBD.8-13 Specifically, co-morbid A? and tau neuropathology, APOE ?4 genotype, lower CSF A?1-42 levels, and increasing A? amyloid burden on PET imaging are all associated with CI in PD. 2) Establishing that specific plasma biomarkers are predictors of CI in PD and AD.14-16 3) Comparing the differential effects of genetic factors on cognition in PD and AD.17-19 4) Other collaborative genetic17, 18, 20-34 and biofluid biomarker studies15, 35-52 demonstrate that SNPs and biofluid biomarkers changes can be disease specific or are altered in many ND. However, we have focused mainly on AD, PD and FTD in our Penn cohorts that enable comparisons of these UPenn data with data from ADNI subjects. Notably, we continue to expand our collaborations with the Blennow/Zetterberg group so that as ADNI Add-On studies identify potentially informative biomarkers in ADNI biosamples going forward, we will follow up with the interrogation of biosamples from the UPenn AD, PD, FTD and ALS subjects. Finally, it shoult be noted that our UPenn cohort is more diverse than the ADNI cohort and we have followed ~150 patients with antemortem biofluid draws to autopsy so we can extend the type of study we reported in Toledo et al.39 to our collaborations with the Blennow/Zetterberg group. This is exemplified in the Cullen et al collaborative study with the Blennow and Zetterberg group (In review , 2018) wherein we collected a panel of five CSF biomarkers ” A?42, total tau (t-tau), phosphorylated tau (p-tau), neurofilament light (NfL), and neurogranin (Ng) ” in 723 subjects representing nine neurodegenerative disorders including both an autopsy-confirmed AD cohort and a clinically representative, probable AD cohort. To seek to differentiate between AD and each of the other disorders in our study cohort, we identified univariate cutoff values of the most informative single biomarker and also fit a multivariate machine learning model using all biomarkers at once. As a result, we found that the multivariate predictive model achieved high differential diagnostic accuracy and greatly outperformed the univariate cutoff model. The strong results on both a gold standard, autopsy-confirmed AD cohort and a clinically representative AD cohort support the idea that this panel is capable of greatly improving patient selection in AD clinical trials.
REFERENCES
1) Robinson JL, Lee EB, Xie SX, Rennert L, Suh E, Bredenberg C, Caswell C, Van Deerlin VM, Yan N, Yousef A, Hurtig HI, Siderowf A, Grossman M, McMillan CT, Miller B, Duda JE, Irwin DJ, Wolk D, Elman L, McCluskey L, Chen-Plotkin A, Weintraub D, Arnold SE, Brettschneider J, Lee VM, Trojanowski JQ. Neurodegenerative disease concomitant proteinopathies are prevalent, age-related and APOE4-associated. Brain;10.1093/brain/awy146, 2018.
2) Kovacs GG, Ferrer I, Grinberg LT, Alafuzoff I, Attems J, Budka H, Cairns NJ, Crary JF, Duyckaerts C, Ghetti B, Halliday GM, Ironside JW, Love S, Mackenzie IR, Munoz DG, Murray ME, Nelson PT, Takahashi H, Trojanowski JQ, Ansorge O, Arzberger T, Baborie A, Beach TG, Bieniek KF, Bigio EH, Bodi I, Dugger BN, Feany M, Gelpi E, Gentleman SM, Giaccone G, Hatanpaa KJ, Heale R, Hof PR, Hofer M, Hortobagyi T, Jellinger K, Jicha GA, Ince P, Kofler J, Kovari E, Kril JJ, Mann DM, Matej R, McKee AC, McLean C, Milenkovic I, Montine TJ, Murayama S, Lee EB, Rahimi J, Rodriguez RD, Rozemuller A, Schneider JA, Schultz C, Seeley W, Seilhean D, Smith C, Tagliavini F, Takao M, Thal DR, Toledo JB, Tolnay M, Troncoso JC, Vinters HV, Weis S, Wharton SB, White CL, 3rd, Wisniewski T, Woulfe JM, Yamada M, Dickson DW. Aging-related tau astrogliopathy (ARTAG): harmonized evaluation strategy. Acta Neuropathol 131(1):87-102, 2016; PMCID: PMC4879001.
3) Kovacs GG, Lee VM, Trojanowski JQ. Protein astrogliopathies in human neurodegenerative diseases and aging. Brain Pathol 27(5):675-90, 2017; PMCID: PMC5578412.
4) Kovacs GG, Robinson JL, Xie SX, Lee EB, Grossman M, Wolk DA, Irwin DJ, Weintraub D, Kim CF, Schuck T, Yousef A, Wagner ST, Suh E, Van Deerlin VM, Lee VM, Trojanowski JQ. Evaluating the Patterns of Aging-Related Tau Astrogliopathy Unravels Novel Insights Into Brain Aging and Neurodegenerative Diseases. J Neuropathol Exp Neurol 76(4):270-88, 2017.
5) Crary JF, Trojanowski JQ, Schneider JA, Abisambra JF, Abner EL, Alafuzoff I, Arnold SE, Attems J, Beach TG, Bigio EH, Cairns NJ, Dickson DW, Gearing M, Grinberg LT, Hof PR, Hyman BT, Jellinger K, Jicha GA, Kovacs GG, Knopman DS, Kofler J, Kukull WA, Mackenzie IR, Masliah E, McKee A, Montine TJ, Murray ME, Neltner JH, Santa-Maria I, Seeley WW, Serrano-Pozo A, Shelanski ML, Stein T, Takao M, Thal DR, Toledo JB, Troncoso JC, Vonsattel JP, White CL, 3rd, Wisniewski T, Woltjer RL, Yamada M, Nelson PT. Primary age-related tauopathy (PART): a common pathology associated with human aging. Acta Neuropathol 128(6):755-66, 2014; PMCID: PMC4257842.
6) Toledo JB, Van Deerlin VM, Lee EB, Suh E, Baek Y, Robinson JL, Xie SX, McBride J, Wood EM, Schuck T, Irwin DJ, Gross RG, Hurtig H, McCluskey L, Elman L, Karlawish J, Schellenberg G, Chen-Plotkin A, Wolk D, Grossman M, Arnold SE, Shaw LM, Lee VM, Trojanowski JQ. A platform for discovery: The University of Pennsylvania Integrated Neurodegenerative Disease Biobank. Alzheimer’s Dement 10(4):477-84.e1, 2014; PMCID: PMC3933464.
7) Arnold SE, Toledo JB, Appleby DH, Xie SX, Wang LS, Baek Y, Wolk DA, Lee EB, Miller BL, Lee VM, Trojanowski JQ. Comparative survey of the topographical distribution of signature molecular lesions in major neurodegenerative diseases. J Comp Neurol 521(18):4339-55, 2013; PMCID: PMC3872132.
8) Akhtar RS, Xie SX, Brennan L, Pontecorvo MJ, Hurtig HI, Trojanowski JQ, Weintraub D, Siderowf AD. Amyloid-Beta Positron Emission Tomography Imaging of Alzheimer’s Pathology in Parkinson’s Disease Dementia. Movement disorders clinical practice 3(4):367-75, 2016; PMCID: PMC4971540.
9) Akhtar RS, Xie SX, Chen YJ, Rick J, Gross RG, Nasrallah IM, Van Deerlin VM, Trojanowski JQ, Chen-Plotkin AS, Hurtig HI, Siderowf AD, Dubroff JG, Weintraub D. Regional brain amyloid-beta accumulation associates with domain-specific cognitive performance in Parkinson disease without dementia. PLoS One 12(5):e0177924, 2017; PMCID: PMC5444629.
10) Irwin DJ, Grossman M, Weintraub D, Hurtig HI, Duda JE, Xie SX, Lee EB, Van Deerlin VM, Lopez OL, Kofler JK, Nelson PT, Jicha GA, Woltjer R, Quinn JF, Kaye J, Leverenz JB, Tsuang D, Longfellow K, Yearout D, Kukull W, Keene CD, Montine TJ, Zabetian CP, Trojanowski JQ. Neuropathological and genetic correlates of survival and dementia onset in synucleinopathies: a retrospective analysis. Lancet Neurol 16(1):55-65, 2017; PMCID: PMC5181646.
11) Simuni T, Siderowf A, Lasch S, Coffey CS, Caspell-Garcia C, Jennings D, Tanner CM, Trojanowski JQ, Shaw LM, Seibyl J, Schuff N, Singleton A, Kieburtz K, Toga AW, Mollenhauer B, Galasko D, Chahine LM, Weintraub D, Foroud T, Tosun D, Poston K, Arnedo V, Frasier M, Sherer T, Chowdhury S, Marek K. Longitudinal change of clinical and biological measures in early Parkinson’s disease: Parkinson’s progression markers initiative cohort. Mov Disord;10.1002/mds.27361, 2018.
12) Shi M, Tang L, Toledo JB, Ginghina C, Wang H, Aro P, Jensen PH, Weintraub D, Chen-Plotkin AS, Irwin DJ, Grossman M, McCluskey L, Elman LB, Wolk DA, Lee EB, Shaw LM, Trojanowski JQ, Zhang J. Cerebrospinal fluid alpha-synuclein contributes to the differential diagnosis of Alzheimer’s disease. Alzheimers Dement;10.1016/j.jalz.2018.02.015, 2018.
13) Irwin DJ, Xie SX, Coughlin D, Nevler N, Akhtar RS, McMillan CT, Lee EB, Wolk DA, Weintraub D, Chen-Plotkin A, Duda JE, Spindler M, Siderowf A, Hurtig HI, Shaw LM, Grossman M, Trojanowski JQ. CSF tau and beta-amyloid predict cerebral synucleinopathy in autopsied Lewy body disorders. Neurology 90(12):e1038-e46, 2018; PMCID: PMC5874449.
14) Lim NS, Swanson CR, Cherng HR, Unger TL, Xie SX, Weintraub D, Marek K, Stern MB, Siderowf A, Trojanowski JQ, Chen-Plotkin AS. Plasma EGF and cognitive decline in Parkinson’s disease and Alzheimer’s disease. Annals of clinical and translational neurology 3(5):346-55, 2016; PMCID: PMC4863747.
15) Berlyand Y, Weintraub D, Xie SX, Mellis IA, Doshi J, Rick J, McBride J, Davatzikos C, Shaw LM, Hurtig H, Trojanowski JQ, Chen-Plotkin AS. An Alzheimer’s Disease-Derived Biomarker Signature Identifies Parkinson’s Disease Patients with Dementia. PLoS One 11(1):e0147319, 2016; PMCID: PMC4727929.
16) Chahine LM, Stern MB, Chen-Plotkin A. Blood-based biomarkers for Parkinson’s disease. Parkinsonism Relat Disord 20 Suppl 1:S99-103, 2014; PMCID: PMC4070332.
17) Mata IF, Leverenz JB, Weintraub D, Trojanowski JQ, Hurtig HI, Van Deerlin VM, Ritz B, Rausch R, Rhodes SL, Factor SA, Wood-Siverio C, Quinn JF, Chung KA, Peterson AL, Espay AJ, Revilla FJ, Devoto J, Hu SC, Cholerton BA, Wan JY, Montine TJ, Edwards KL, Zabetian CP. APOE, MAPT, and SNCA genes and cognitive performance in Parkinson disease. JAMA neurology 71(11):1405-12, 2014; PMCID: PMC4227942.
18) Mata IF, Leverenz JB, Weintraub D, Trojanowski JQ, Chen-Plotkin A, Van Deerlin VM, Ritz B, Rausch R, Factor SA, Wood-Siverio C, Quinn JF, Chung KA, Peterson-Hiller AL, Goldman JG, Stebbins GT, Bernard B, Espay AJ, Revilla FJ, Devoto J, Rosenthal LS, Dawson TM, Albert MS, Tsuang D, Huston H, Yearout D, Hu SC, Cholerton BA, Montine TJ, Edwards KL, Zabetian CP. GBA Variants are associated with a distinct pattern of cognitive deficits in Parkinson’s disease. Mov Disord 31(1):95-102, 2016; PMCID: PMC4724255.
19) Mata IF, Johnson CO, Leverenz JB, Weintraub D, Trojanowski JQ, Van Deerlin VM, Ritz B, Rausch R, Factor SA, Wood-Siverio C, Quinn JF, Chung KA, Peterson-Hiller AL, Espay AJ, Revilla FJ, Devoto J, Yearout D, Hu SC, Cholerton BA, Montine TJ, Edwards KL, Zabetian CP. Large-scale exploratory genetic analysis of cognitive impairment in Parkinson’s disease. Neurobiol Aging 56:211.e1-.e7, 2017; PMCID: PMC5536182.
20) Beecham GW, Hamilton K, Naj AC, Martin ER, Huentelman M, Myers AJ, Corneveaux JJ, Hardy J, Vonsattel JP, Younkin SG, Bennett DA, De Jager PL, Larson EB, Crane PK, Kamboh MI, Kofler JK, Mash DC, Duque L, Gilbert JR, Gwirtsman H, Buxbaum JD, Kramer P, Dickson DW, Farrer LA, Frosch MP, Ghetti B, Haines JL, Hyman BT, Kukull WA, Mayeux RP, Pericak-Vance MA, Schneider JA, Trojanowski JQ, Reiman EM, Alzheimer’s Disease Genetics C, Schellenberg GD, Montine TJ. Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS Genet 10(9):e1004606, 2014; PMCID: PMC4154667.
21) Beecham GW, Dickson DW, Scott WK, Martin ER, Schellenberg G, Nuytemans K, Larson EB, Buxbaum JD, Trojanowski JQ, Van Deerlin VM, Hurtig HI, Mash DC, Beach TG, Troncoso JC, Pletnikova O, Frosch MP, Ghetti B, Foroud TM, Honig LS, Marder K, Vonsattel JP, Goldman SM, Vinters HV, Ross OA, Wszolek ZK, Wang L, Dykxhoorn DM, Pericak-Vance MA, Montine TJ, Leverenz JB, Dawson TM, Vance JM. PARK10 is a major locus for sporadic neuropathologically confirmed Parkinson disease. Neurology 84(10):972-80, 2015; PMCID: PMC4352096.
22) Chahine LM, Qiang J, Ashbridge E, Minger J, Yearout D, Horn S, Colcher A, Hurtig HI, Lee VM, Van Deerlin VM, Leverenz JB, Siderowf AD, Trojanowski JQ, Zabetian CP, Chen-Plotkin A. Clinical and biochemical differences in patients having Parkinson disease with vs without GBA mutations. JAMA neurology 70(7):852-8, 2013; PMCID: PMC3762458.
23) Kalia LV, Lang AE, Hazrati LN, Fujioka S, Wszolek ZK, Dickson DW, Ross OA, Van Deerlin VM, Trojanowski JQ, Hurtig HI, Alcalay RN, Marder KS, Clark LN, Gaig C, Tolosa E, Ruiz-Martinez J, Marti-Masso JF, Ferrer I, Lopez de Munain A, Goldman SM, Schule B, Langston JW, Aasly JO, Giordana MT, Bonifati V, Puschmann A, Canesi M, Pezzoli G, Maues De Paula A, Hasegawa K, Duyckaerts C, Brice A, Stoessl AJ, Marras C. Clinical correlations with Lewy body pathology in LRRK2-related Parkinson disease. JAMA neurology 72(1):100-5, 2015; PMCID: PMC4399368.
24) Lill CM, Rengmark A, Pihlstrom L, Fogh I, Shatunov A, Sleiman PM, Wang LS, Liu T, Lassen CF, Meissner E, Alexopoulos P, Calvo A, Chio A, Dizdar N, Faltraco F, Forsgren L, Kirchheiner J, Kurz A, Larsen JP, Liebsch M, Linder J, Morrison KE, Nissbrandt H, Otto M, Pahnke J, Partch A, Restagno G, Rujescu D, Schnack C, Shaw CE, Shaw PJ, Tumani H, Tysnes OB, Valladares O, Silani V, van den Berg LH, van Rheenen W, Veldink JH, Lindenberger U, Steinhagen-Thiessen E, Consortium S, Teipel S, Perneczky R, Hakonarson H, Hampel H, von Arnim CA, Olsen JH, Van Deerlin VM, Al-Chalabi A, Toft M, Ritz B, Bertram L. The role of TREM2 R47H as a risk factor for Alzheimer’s disease, frontotemporal lobar degeneration, amyotrophic lateral sclerosis, and Parkinson’s disease. Alzheimers Dement;10.1016/j.jalz.2014.12.009, 2015.
25) Mata IF, Checkoway H, Hutter CM, Samii A, Roberts JW, Kim HM, Agarwal P, Alvarez V, Ribacoba R, Pastor P, Lorenzo-Betancor O, Infante J, Sierra M, Gomez-Garre P, Mir P, Ritz B, Rhodes SL, Colcher A, Van Deerlin V, Chung KA, Quinn JF, Yearout D, Martinez E, Farin FM, Wan JY, Edwards KL, Zabetian CP. Common variation in the LRRK2 gene is a risk factor for Parkinson’s disease. Mov Disord 27(14):1822-5, 2012; PMCID: PMC3536918.
26) Nalls MA, McLean CY, Rick J, Eberly S, Hutten SJ, Gwinn K, Sutherland M, Martinez M, Heutink P, Williams NM, Hardy J, Gasser T, Brice A, Price TR, Nicolas A, Keller MF, Molony C, Gibbs JR, Chen-Plotkin A, Suh E, Letson C, Fiandaca MS, Mapstone M, Federoff HJ, Noyce AJ, Morris H, Van Deerlin VM, Weintraub D, Zabetian C, Hernandez DG, Lesage S, Mullins M, Conley ED, Northover CA, Frasier M, Marek K, Day-Williams AG, Stone DJ, Ioannidis JP, Singleton AB. Diagnosis of Parkinson’s disease on the basis of clinical and genetic classification: a population-based modelling study. Lancet Neurol 14(10):1002-9, 2015; PMCID: PMC4575273.
27) Nalls MA, Duran R, Lopez G, Kurzawa-Akanbi M, McKeith IG, Chinnery PF, Morris CM, Theuns J, Crosiers D, Cras P, Engelborghs S, De Deyn PP, Van Broeckhoven C, Mann DM, Snowden J, Pickering-Brown S, Halliwell N, Davidson Y, Gibbons L, Harris J, Sheerin UM, Bras J, Hardy J, Clark L, Marder K, Honig LS, Berg D, Maetzler W, Brockmann K, Gasser T, Novellino F, Quattrone A, Annesi G, De Marco EV, Rogaeva E, Masellis M, Black SE, Bilbao JM, Foroud T, Ghetti B, Nichols WC, Pankratz N, Halliday G, Lesage S, Klebe S, Durr A, Duyckaerts C, Brice A, Giasson BI, Trojanowski JQ, Hurtig HI, Tayebi N, Landazabal C, Knight MA, Keller M, Singleton AB, Wolfsberg TG, Sidransky E. A multicenter study of glucocerebrosidase mutations in dementia with Lewy bodies. JAMA neurology 70(6):727-35, 2013; PMCID: PMC3841974.
28) Nuytemans K, Inchausti V, Beecham GW, Wang L, Dickson DW, Trojanowski JQ, Lee VM, Mash DC, Frosch MP, Foroud TM, Honig LS, Montine TJ, Dawson TM, Martin ER, Scott WK, Vance JM. Absence of C9ORF72 expanded or intermediate repeats in autopsy-confirmed Parkinson’s disease. Mov Disord 29(6):827-30, 2014; PMCID: PMC4022044.
29) Pankratz N, Beecham GW, DeStefano AL, Dawson TM, Doheny KF, Factor SA, Hamza TH, Hung AY, Hyman BT, Ivinson AJ, Krainc D, Latourelle JC, Clark LN, Marder K, Martin ER, Mayeux R, Ross OA, Scherzer CR, Simon DK, Tanner C, Vance JM, Wszolek ZK, Zabetian CP, Myers RH, Payami H, Scott WK, Foroud T. Meta-analysis of Parkinson’s disease: identification of a novel locus, RIT2. Ann Neurol 71(3):370-84, 2012; PMCID: PMC3354734.
30) Tsuang D, Leverenz JB, Lopez OL, Hamilton RL, Bennett DA, Schneider JA, Buchman AS, Larson EB, Crane PK, Kaye JA, Kramer P, Woltjer R, Kukull W, Nelson PT, Jicha GA, Neltner JH, Galasko D, Masliah E, Trojanowski JQ, Schellenberg GD, Yearout D, Huston H, Fritts-Penniman A, Mata IF, Wan JY, Edwards KL, Montine TJ, Zabetian CP. GBA mutations increase risk for Lewy body disease with and without Alzheimer disease pathology. Neurology 79(19):1944-50, 2012; PMCID: PMC3484986.
31) Tsuang D, Leverenz JB, Lopez OL, Hamilton RL, Bennett DA, Schneider JA, Buchman AS, Larson EB, Crane PK, Kaye JA, Kramer P, Woltjer R, Trojanowski JQ, Weintraub D, Chen-Plotkin AS, Irwin DJ, Rick J, Schellenberg GD, Watson GS, Kukull W, Nelson PT, Jicha GA, Neltner JH, Galasko D, Masliah E, Quinn JF, Chung KA, Yearout D, Mata IF, Wan JY, Edwards KL, Montine TJ, Zabetian CP. APOE epsilon4 increases risk for dementia in pure synucleinopathies. JAMA neurology 70(2):223-8, 2013; PMCID: PMC3580799.
32) Kun-Rodrigues C, Ross OA, Orme T, Shepherd C, Parkkinen L, Darwent L, Hernandez D, Ansorge O, Clark LN, Honig LS, Marder K, Lemstra A, Scheltens P, van der Flier W, Louwersheimer E, Holstege H, Rogaeva E, St George-Hyslop P, Londos E, Zetterberg H, Barber I, Braae A, Brown K, Morgan K, Maetzler W, Berg D, Troakes C, Al-Sarraj S, Lashley T, Holton J, Compta Y, Van Deerlin V, Trojanowski JQ, Serrano GE, Beach TG, Clarimon J, Lleo A, Morenas-Rodriguez E, Lesage S, Galasko D, Masliah E, Santana I, Diez M, Pastor P, Tienari PJ, Myllykangas L, Oinas M, Revesz T, Lees A, Boeve BF, Petersen RC, Ferman TJ, Escott-Price V, Graff-Radford N, Cairns NJ, Morris JC, Stone DJ, Pickering-Brown S, Mann D, Dickson DW, Halliday GM, Singleton A, Guerreiro R, Bras J. Analysis of C9orf72 repeat expansions in a large international cohort of dementia with Lewy bodies. Neurobiol Aging;10.1016/j.neurobiolaging.2016.08.023, 2016.
33) Wang LS, Naj AC, Graham RR, Crane PK, Kunkle BW, Cruchaga C, Murcia JD, Cannon-Albright L, Baldwin CT, Zetterberg H, Blennow K, Kukull WA, Faber KM, Schupf N, Norton MC, Tschanz JT, Munger RG, Corcoran CD, Rogaeva E, Alzheimer’s Disease Genetics C, Lin CF, Dombroski BA, Cantwell LB, Partch A, Valladares O, Hakonarson H, St George-Hyslop P, Green RC, Goate AM, Foroud TM, Carney RM, Larson EB, Behrens TW, Kauwe JS, Haines JL, Farrer LA, Pericak-Vance MA, Mayeux R, Schellenberg GD, National Institute on Aging-Late-Onset Alzheimer’s Disease Family S, Albert MS, Albin RL, Apostolova LG, Arnold SE, Barber R, Barmada M, Barnes LL, Beach TG, Becker JT, Beecham GW, Beekly D, Bennett DA, Bigio EH, Bird TD, Blacker D, Boeve BF, Bowen JD, Boxer A, Burke JR, Buxbaum JD, Cairns NJ, Cao C, Carlson CS, Carroll SL, Chui HC, Clark DG, Cribbs DH, Crocco EA, DeCarli C, DeKosky ST, Demirci FY, Dick M, Dickson DW, Duara R, Ertekin-Taner N, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Glass JD, Graff-Radford NR, Growdon JH, Hamilton RL, Hamilton-Nelson KL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jarvik GP, Jicha GA, Jin LW, Jun G, Jun G, Kamboh MI, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Kramer JH, LaFerla FM, Lah JJ, Leverenz JB, Levey AI, Li G, Lieberman AP, Lopez OL, Lunetta KL, Lyketsos CG, Mack WJ, Marson DC, Martin ER, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam WM, Miller BL, Miller CA, Miller JW, Montine TJ, Morris JC, Murrell JR, Olichney JM, Parisi JE, Perry W, Peskind E, Petersen RC, Pierce A, Poon WW, Potter H, Quinn JF, Raj A, Raskind M, Reiman EM, Reisberg B, Reitz C, Ringman JM, Roberson ED, Rosen HJ, Rosenberg RN, Sano M, Saykin AJ, Schneider JA, Schneider LS, Seeley WW, Smith AG, Sonnen JA, Spina S, Stern RA, Tanzi RE, Thornton-Wells TA, Trojanowski JQ, Troncoso JC, Tsuang DW, Van Deerlin VM, Van Eldik LJ, Vardarajan BN, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Wishnek S, Woltjer RL, Wright CB, Younkin SG, Yu CE, Yu L. Rarity of the Alzheimer disease-protective APP A673T variant in the United States. JAMA neurology 72(2):209-16, 2015; PMCID: PMC4324097.
34) McMillan CT, Toledo JB, Avants BB, Cook PA, Wood EM, Suh E, Irwin DJ, Powers J, Olm C, Elman L, McCluskey L, Schellenberg GD, Lee VM, Trojanowski JQ, Van Deerlin VM, Grossman M. Genetic and neuroanatomic associations in sporadic frontotemporal lobar degeneration. Neurobiol Aging 35(6):1473-82, 2014; PMCID: PMC3961542.
35) Kang JH, Irwin DJ, Chen-Plotkin AS, Siderowf A, Caspell C, Coffey CS, Waligorska T, Taylor P, Pan S, Frasier M, Marek K, Kieburtz K, Jennings D, Simuni T, Tanner CM, Singleton A, Toga AW, Chowdhury S, Mollenhauer B, Trojanowski JQ, Shaw LM, Parkinson’s Progression Markers I. Association of cerebrospinal fluid beta-amyloid 1-42, T-tau, P-tau181, and alpha-synuclein levels with clinical features of drug-naive patients with early Parkinson disease. JAMA neurology 70(10):1277-87, 2013; PMCID: PMC4034348.
36) Kang JH, Mollenhauer B, Coffey CS, Toledo JB, Weintraub D, Galasko DR, Irwin DJ, Van Deerlin V, Chen-Plotkin AS, Caspell-Garcia C, Waligorska T, Taylor P, Shah N, Pan S, Zero P, Frasier M, Marek K, Kieburtz K, Jennings D, Tanner CM, Simuni T, Singleton A, Toga AW, Chowdhury S, Trojanowski JQ, Shaw LM, Parkinson’s Progression Marker I. CSF biomarkers associated with disease heterogeneity in early Parkinson’s disease: the Parkinson’s Progression Markers Initiative study. Acta Neuropathol 131(6):935-49, 2016.
37) Lim NS, Swanson CR, Cherng HR, Unger TL, Xie SX, Weintraub D, Marek K, Stern MB, Siderowf A, Investigators P, Alzheimer’s Disease Neuroimaging I, Trojanowski JQ, Chen-Plotkin AS. Plasma EGF and cognitive decline in Parkinson’s disease and Alzheimer’s disease. Annals of clinical and translational neurology 3(5):346-55, 2016; PMCID: PMC4863747.
38) Swanson CR, Li K, Unger TL, Gallagher MD, Van Deerlin VM, Agarwal P, Leverenz J, Roberts J, Samii A, Gross RG, Hurtig H, Rick J, Weintraub D, Trojanowski JQ, Zabetian C, Chen-Plotkin AS. Lower plasma apolipoprotein A1 levels are found in Parkinson’s disease and associate with apolipoprotein A1 genotype. Mov Disord 30(6):805-12, 2015; PMCID: PMC4362847.
39) Toledo JB, Brettschneider J, Grossman M, Arnold SE, Hu WT, Xie SX, Lee VM, Shaw LM, Trojanowski JQ. CSF biomarkers cutoffs: the importance of coincident neuropathological diseases. Acta Neuropathol 124(1):23-35, 2012; PMCID: PMC3551449.
40) Toledo JB, Korff A, Shaw LM, Trojanowski JQ, Zhang J. CSF alpha-synuclein improves diagnostic and prognostic performance of CSF tau and Abeta in Alzheimer’s disease. Acta Neuropathol 126(5):683-97, 2013; PMCID: PMC3812407.
41) Swanson CR, Berlyand Y, Xie SX, Alcalay RN, Chahine LM, Chen-Plotkin AS. Plasma apolipoprotein A1 associates with age at onset and motor severity in early Parkinson’s disease patients. Mov Disord 30(12):1648-56, 2015; PMCID: PMC4609229.
42) Portelius E, Olsson B, Hoglund K, Cullen NC, Kvartsberg H, Andreasson U, Zetterberg H, Sandelius A, Shaw LM, Lee VMY, Irwin DJ, Grossman M, Weintraub D, Chen-Plotkin A, Wolk DA, McCluskey L, Elman L, McBride J, Toledo JB, Trojanowski JQ, Blennow K. Cerebrospinal fluid neurogranin concentration in neurodegeneration: relation to clinical phenotypes and neuropathology. Acta Neuropathol;10.1007/s00401-018-1851-x, 2018.
43) O’Bryant SE, Gupta V, Henriksen K, Edwards M, Jeromin A, Lista S, Bazenet C, Soares H, Lovestone S, Hampel H, Montine T, Blennow K, Foroud T, Carrillo M, Graff-Radford N, Laske C, Breteler M, Shaw L, Trojanowski JQ, Schupf N, Rissman RA, Fagan AM, Oberoi P, Umek R, Weiner MW, Grammas P, Posner H, Martins R, Star B, groups Bw. Guidelines for the standardization of preanalytic variables for blood-based biomarker studies in Alzheimer’s disease research. Alzheimers Dement 11(5):549-60, 2015; PMCID: PMC4414664.
44) Lewczuk P, Riederer P, O’Bryant SE, Verbeek MM, Dubois B, Visser PJ, Jellinger KA, Engelborghs S, Ramirez A, Parnetti L, Jack CR, Jr., Teunissen CE, Hampel H, Lleo A, Jessen F, Glodzik L, de Leon MJ, Fagan AM, Molinuevo JL, Jansen WJ, Winblad B, Shaw LM, Andreasson U, Otto M, Mollenhauer B, Wiltfang J, Turner MR, Zerr I, Handels R, Thompson AG, Johansson G, Ermann N, Trojanowski JQ, Karaca I, Wagner H, Oeckl P, van Waalwijk van Doorn L, Bjerke M, Kapogiannis D, Kuiperij HB, Farotti L, Li Y, Gordon BA, Epelbaum S, Vos SJB, Klijn CJM, Van Nostrand WE, Minguillon C, Schmitz M, Gallo C, Lopez Mato A, Thibaut F, Lista S, Alcolea D, Zetterberg H, Blennow K, Kornhuber J. Cerebrospinal fluid and blood biomarkers for neurodegenerative dementias: An update of the Consensus of the Task Force on Biological Markers in Psychiatry of the World Federation of Societies of Biological Psychiatry. World J Biol Psychiatry 19(4):244-328, 2018; PMCID: PMC5916324.
45) Henriksen K, O’Bryant SE, Hampel H, Trojanowski JQ, Montine TJ, Jeromin A, Blennow K, Lonneborg A, Wyss-Coray T, Soares H, Bazenet C, Sjogren M, Hu W, Lovestone S, Karsdal MA, Weiner MW, Blood-Based Biomarker Interest G. The future of blood-based biomarkers for Alzheimer’s disease. Alzheimers Dement 10(1):115-31, 2014; PMCID: PMC4128378.
46) Mattsson N, Andreasson U, Persson S, Carrillo MC, Collins S, Chalbot S, Cutler N, Dufour-Rainfray D, Fagan AM, Heegaard NH, Robin Hsiung GY, Hyman B, Iqbal K, Kaeser SA, Lachno DR, Lleo A, Lewczuk P, Molinuevo JL, Parchi P, Regeniter A, Rissman RA, Rosenmann H, Sancesario G, Schroder J, Shaw LM, Teunissen CE, Trojanowski JQ, Vanderstichele H, Vandijck M, Verbeek MM, Zetterberg H, Blennow K. CSF biomarker variability in the Alzheimer’s Association quality control program. Alzheimers Dement 9(3):251-61, 2013; PMCID: PMC3707386.
47) Toledo JB, Shaw LM, Trojanowski JQ. Plasma amyloid beta measurements – a desired but elusive Alzheimer’s disease biomarker. Alzheimer’s research & therapy 5(2):8, 2013; PMCID: PMC3706955.
48) Toledo JB, Korff A, Shaw LM, Trojanowski JQ, Zhang J. Low levels of cerebrospinal fluid complement 3 and factor H predict faster cognitive decline in mild cognitive impairment. Alzheimer’s research & therapy 6(3):36, 2014; PMCID: Pmc4255518.
49) McMillan CT, Avants B, Irwin DJ, Toledo JB, Wolk DA, Van Deerlin VM, Shaw LM, Trojanoswki JQ, Grossman M. Can MRI screen for CSF biomarkers in neurodegenerative disease? Neurology 80(2):132-8, 2013; PMCID: PMC3589187.
50) Lleo A, Irwin DJ, Illan-Gala I, McMillan CT, Wolk DA, Lee EB, Van Deerlin VM, Shaw LM, Trojanowski JQ, Grossman M. A 2-Step Cerebrospinal Algorithm for the Selection of Frontotemporal Lobar Degeneration Subtypes. JAMA neurology 75(6):738-45, 2018; PMCID: PMC5885205.
51) Spotorno N, McMillan CT, Irwin DJ, Clark R, Lee EB, Trojanowski JQ, Weintraub D, Grossman M. Decision-Making Deficits Associated with Amyloidosis in Lewy Body Disorders. Front Hum Neurosci 10:693, 2016; PMCID: PMC5225123.
52) Grossman M, Elman L, McCluskey L, McMillan CT, Boller A, Powers J, Rascovsky K, Hu W, Shaw L, Irwin DJ, Lee VM, Trojanowski JQ. Phosphorylated tau as a candidate biomarker for amyotrophic lateral sclerosis. JAMA neurology 71(4):442-8, 2014; PMCID: PMC3989393.
Please refer to the following manuscripts for comparison with ADNI data files: